Basic local alignment search tool pdf Manawatu-Wanganui

(PDF) Basic local alignment search tool loveneet singh Basic Local Alignment Search Tool(BLAST) L39 5 2. The higher the percentage of hits obtained indicates the more likely of the query related to the matches. But, if the similarity is only in small regions, this likely indicates related functional domains rather than related proteins/gene products.
Foreword by B LA ST pedagogix-tagc.univ-mrs.fr
HBLAST Parallelised sequence similarity – A Hadoop. 16/07/2016 · (2008) Blast (basic local alignment search tool). In: Encyclopedia of Genetics, Genomics, Proteomics and Informatics. Springer, Dordrecht.RIS Papers …, 09/11/2017 · In this video, we describe the conceptual background and analysis method of Nucleotide BLAST (Basic Local Alignment Search Tool, BLASTn) analysis. Video on Local and Global Alignment ….
31/05/2019В В· Download BLAST Basic Local Alignment Search Tool book pdf free download link or read online here in PDF. Read online BLAST Basic Local Alignment Search Tool book pdf free download link book now. All books are in clear copy here, and all files are secure so don't worry about it. This site is like a library, you could find million book here by Using the Basic Local Alignment Search Tool (BLAST) FASTA finds short common patterns in query and database sequences and joins these into an alignment. BLAST is similar to FASTA, but gains a further increase in speed by searching only for rarer, more significant patterns in nucleic acid and protein sequences. BLAST is very popular due to its availability on the World Wide Web through a
BLAST (acronyme de basic local alignment search tool) est une mГ©thode de recherche heuristique utilisГ©e en bio-informatique. Il permet de trouver les rГ©gions similaires entre deux ou plusieurs sГ©quences de nuclГ©otides ou d'acides aminГ©s, et de rГ©aliser un alignement de ces rГ©gions homologues. The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches. BLAST can be used to infer functional and evolutionary relationships between sequences as well as help identify
The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches. BLAST can be used to infer functional and evolutionary relationships between sequences as well as help identify the Basic Local Alignment Search Tool Ian Korf, Mark Yandell & Joseph Bedell B LA ST Foreword by Stephen Altschul. BLAST. Other resources from OГ•Reilly oreilly.com oreilly.com is more than a complete catalog of O'Reilly books. You'll also find links to news, events, articles, weblogs, sample chapters, and code examples. oreillynet.com is the essential portal for developers interested in open
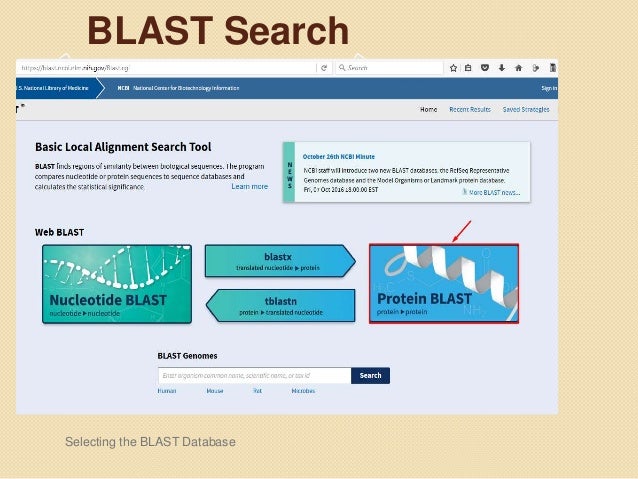
Basic Local Alignment Search Tool (BLAST) BLAST is a popular user-friendly tool for searching all the major sequence databases.BLAST is a heuristic method to find the highest scoring locally optimal alignments between a query sequence and a database sequence. BLAST programs of sequences, making it a critical tool to ongoing genomic research. In fact, the initial paper describing the program, titled "Basic Local Alignment Search Tool" and published in the Journal of Molecular Biology, was the most highly cited publication of the 1990s (Taubs, 2000). The parallel
Recherche par similaritГ© dans les banques/bases de donnГ©es La suite Blast (Basic Local Alignment Search Tool) 31/05/2019В В· Download BLAST Basic Local Alignment Search Tool book pdf free download link or read online here in PDF. Read online BLAST Basic Local Alignment Search Tool book pdf free download link book now. All books are in clear copy here, and all files are secure so don't worry about it. This site is like a library, you could find million book here by
search –Lower the E-value higher is the significance of score –P-value indicates if such an alignment can be expected from a chance alone Chance: can mean the comparison of (a) real but non-homologous sequences (True negatives) (b) real sequences that are shuffled to preserve compositional properties (c) sequences that are generated randomly The Basic Local Alignment Search Tool (BLAST) , a heuristic version of the pairwise local alignment Smith Waterman algorithm , remains the most widely used computational procedure for alignment interrogating biological databases based on a heuristic version of the pairwise local alignment Smith Waterman algorithm. It compares the similarity of
BF children showed a significant enrichment in Bacteroidetes and depletion in Firmicutes (P < 0.001), with a unique abundance of bacteria from the genus Prevotella and Xylanibacter, known to contain a set of View Basic Local Alignment Search Tool.pdf from AA 1Basic Local Alignment Search Tool BLAST Why Use BLAST? BLAST helps you to find homologous genes and proteins Homologous Proteins (or genes) •
A new approach to rapid sequence comparison, basic local alignment search tool (BLAST), directly approximates alignments that optimize a measure of local similarity, the maximal segment pair (MSP In this paper we describe a new method, BLAST†(Basic Local Alignment Search Tool), which employs a measure based on wellВdefined mutation scores. It directly approximates the results that would be obtained by a dynamic programming algorithm for optimizing this …
1 BLAST: Basic Local Alignment Search Tool Jonathan M. Urbach Bioinformatics Group Department of Molecular Biology (Basic Local Alignment Search Tool), which employs a measure based on well-defined mutation scores. It directly approximates the results that would be obtained by a dynamic programming algo- rithm for optimizing this measure. The method will detect weak but biologically significant sequence
Created Date: 7/12/2001 9:55:01 AM 29/07/2010В В· Tutorial for BLAST, a cornerstone bioinformatics tool at NCBI. BLAST is the BLAST is the Basic Local Alignment Search tool and will protein and DNA sequences that
Basic Local Alignment Search Tool NDSU

An Essential Guide to the Basic Local Alignment Search. BLAST Basic Local Alignment Search Tool Job Title: lcl|P76387|WZC_ECOLI No comment found Show Conserved Domains Putative conserved domains have been …, 29/07/2010 · Tutorial for BLAST, a cornerstone bioinformatics tool at NCBI. BLAST is the BLAST is the Basic Local Alignment Search tool and will protein and DNA sequences that.
(PDF) Basic local alignment search tool N w Academia.edu
[PDF] Basic Local Alignment Search Tool Scinapse. Basic Local Alignment Search Tool Stephen F. Altschul', Warren Gish', Webb Miller2 Eugene W. Myers3 and David J. Lipmanl 'Satwnal Center for Biotechnology Informalion Kdional Library of … https://fa.wikipedia.org/wiki/%D8%A8%D9%84%D8%A7%D8%B3%D8%AA search –Lower the E-value higher is the significance of score –P-value indicates if such an alignment can be expected from a chance alone Chance: can mean the comparison of (a) real but non-homologous sequences (True negatives) (b) real sequences that are shuffled to preserve compositional properties (c) sequences that are generated randomly.
BLAST (acronyme de basic local alignment search tool) est une mГ©thode de recherche heuristique utilisГ©e en bio-informatique. Il permet de trouver les rГ©gions similaires entre deux ou plusieurs sГ©quences de nuclГ©otides ou d'acides aminГ©s, et de rГ©aliser un alignement de ces rГ©gions homologues. A new approach to rapid sequence comparison, basic local alignment search tool (BLAST), directly approximates alignments that optimize a measure of local similarity, the maximal segment pair (MSP) score. Recent mathematical results on the stochastic properties of MSP scores allow an analysis of the performance of this method as well as the
(Basic Local Alignment Search Tool), which employs a measure based on well-defined mutation scores. It . directly approximates the results that would he obt,ained by a dynamic programming algoВ rithm for optimizing this measure. The method will detect weak but biologically significant sequence Introduction to Basic Local Alignment Search Tool 27 the discovery that the defective gene that caused cystic fibrosis formed a protein that had similarity to a family of proteins involved in the transport of hydrophilic molecules across the cytoplasmic membrane (Riordan, et. al., 1989). Cystic fibrosis is the most common inherited disease in the
BLAST (acronyme de basic local alignment search tool) est une méthode de recherche heuristique utilisée en bio-informatique. Il permet de trouver les régions similaires entre deux ou plusieurs séquences de nucléotides ou d'acides aminés, et de réaliser un alignement de ces régions homologues. Basic Local Alignment Search Tool Stephen F. Altschul', Warren Gish', Webb Miller2 Eugene W. Myers3 and David J. Lipmanl 'Satwnal Center for Biotechnology Informalion Kdional Library of …
CSC 448 Bioinformatics Algorithms Alexander Dekhtyar. . (Rapid) Local Sequence Alignment BLAST BLAST: Basic Local Alignment Search Tool BLASTis afamilyof rapid approximate localalignmentalgorithms[2]. BF children showed a significant enrichment in Bacteroidetes and depletion in Firmicutes (P < 0.001), with a unique abundance of bacteria from the genus Prevotella and Xylanibacter, known to contain a set of
search –Lower the E-value higher is the significance of score –P-value indicates if such an alignment can be expected from a chance alone Chance: can mean the comparison of (a) real but non-homologous sequences (True negatives) (b) real sequences that are shuffled to preserve compositional properties (c) sequences that are generated randomly This manual documents the BLAST (Basic Local Alignment Search Tool) command line applications developed at the National Center for Biotechnology Information (NCBI).
(Basic Local Alignment Search Tool), which employs a measure based on well-defined mutation scores. It . directly approximates the results that would he obt,ained by a dynamic programming algoВ rithm for optimizing this measure. The method will detect weak but biologically significant sequence Introduction to Basic Local Alignment Search Tool 27 the discovery that the defective gene that caused cystic fibrosis formed a protein that had similarity to a family of proteins involved in the transport of hydrophilic molecules across the cytoplasmic membrane (Riordan, et. al., 1989). Cystic fibrosis is the most common inherited disease in the
31/05/2019В В· Download BLAST Basic Local Alignment Search Tool book pdf free download link or read online here in PDF. Read online BLAST Basic Local Alignment Search Tool book pdf free download link book now. All books are in clear copy here, and all files are secure so don't worry about it. This site is like a library, you could find million book here by Wet lab Sampling (fish eye parasites) DNA extraction Primer design/ COI (marker ) PCR Library preparation and NGS (Illumina) Paired End sequencing
(Basic Local Alignment Search Tool), which employs a measure based on well-defined mutation scores. It directly approximates the results that would be obtained by a dynamic programming algo- rithm for optimizing this measure. The method will detect weak but biologically significant sequence The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches. BLAST can be used to infer functional and evolutionary relationships between sequences as well as help identify
BF children showed a significant enrichment in Bacteroidetes and depletion in Firmicutes (P < 0.001), with a unique abundance of bacteria from the genus Prevotella and Xylanibacter, known to contain a set of 29/07/2010В В· Tutorial for BLAST, a cornerstone bioinformatics tool at NCBI. BLAST is the BLAST is the Basic Local Alignment Search tool and will protein and DNA sequences that
29/07/2010 · Tutorial for BLAST, a cornerstone bioinformatics tool at NCBI. BLAST is the BLAST is the Basic Local Alignment Search tool and will protein and DNA sequences that 09/11/2017 · In this video, we describe the conceptual background and analysis method of Nucleotide BLAST (Basic Local Alignment Search Tool, BLASTn) analysis. Video on Local and Global Alignment …
search –Lower the E-value higher is the significance of score –P-value indicates if such an alignment can be expected from a chance alone Chance: can mean the comparison of (a) real but non-homologous sequences (True negatives) (b) real sequences that are shuffled to preserve compositional properties (c) sequences that are generated randomly The Basic Local Alignment Search Tool (BLAST) , a heuristic version of the pairwise local alignment Smith Waterman algorithm , remains the most widely used computational procedure for alignment interrogating biological databases based on a heuristic version of the pairwise local alignment Smith Waterman algorithm. It compares the similarity of
The Cub Cadet LTX 1050 (50-Inch) 24HP Lawn Tractor 13WQ93AP010 has been discontinued. Check out Expert's recommended alternatives for another top lawn mower accessory. Cub cadet ltx 1050 service manual West Coast 07/11/2019В В· I have a Cub Cadet LTX1050 with a 22HP Kohler Courage engine (model SV715. I recently installed the PTO and the spindles on the mower. When I engaged to PTO the battery light came on. I then bought a new battery (over 5 years old) I then checked the regulator and got a reading of 45V on the A+A connectors and -3 DCV on the B connector. I knew
BLAST Basic Local Alignment Search Tool BYKdb

Jonathan M. Urbach Bioinformatics Group Department of. We have compared our result with the approximate solution obtained with BLAST (basic local alignment search tool) software, which is currently the most widely used for searching for protein alignment., BLAST Basic Local Alignment Search Tool Job Title: Borrprobe Your search parameters were adjusted to search for a short input sequence. BLASTN 2.2.17 (Aug-26-2007) Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, ….
NCBI Blast Tutorial YouTube
Command Line Applications User Manual NCBI Bookshelf. Basic Local Alignment Search Tool Martin T otsches 14.06.2010 Martin T otsches Basic Local Alignment Search Tool. History Alignments Scores BLAST algorithm Conclusion Table of contents 1 History 2 Alignments global Alignments local Alignments 3 Scores Score functions Score matrices 4 BLAST algorithm Methods Statistical signi cance Performance 5 Conclusion Comparison to FASTA …, The Basic Local Alignment Search Tool (BLAST) , a heuristic version of the pairwise local alignment Smith Waterman algorithm , remains the most widely used computational procedure for alignment interrogating biological databases based on a heuristic version of the pairwise local alignment Smith Waterman algorithm. It compares the similarity of.
Introduction to Basic Local Alignment Search Tool. Chapter. 6 Readers; 1.3k Downloads; Summary . In this Chapter, we created a Swing based application that allows users to prepare sequences for BLAST searches by performing simple formatting tasks such as conversion into the Fasta format and determining the sequence type and length. Along the way we introduced how to write code to respond … A new approach to rapid sequence comparison, basic local alignment search tool (BLAST), directly approximates alignments that optimize a measure of local similarity, the maximal segment pair (MSP) score. Recent mathematical results on the stochastic properties of MSP scores allow an analysis of the performance of this method as well as the
Basic Local Alignment Search Tool. Alignments • used to uncover homologies between sequences • combined with phylogenetic studies o can determine orthologous and paralogous relationships Introduction to Basic Local Alignment Search Tool. Chapter. 6 Readers; 1.3k Downloads; Summary . In this Chapter, we created a Swing based application that allows users to prepare sequences for BLAST searches by performing simple formatting tasks such as conversion into the Fasta format and determining the sequence type and length. Along the way we introduced how to write code to respond …
This manual documents the BLAST (Basic Local Alignment Search Tool) command line applications developed at the National Center for Biotechnology Information (NCBI). 1 BLAST: Basic Local Alignment Search Tool Jonathan M. Urbach Bioinformatics Group Department of Molecular Biology
Wet lab Sampling (fish eye parasites) DNA extraction Primer design/ COI (marker ) PCR Library preparation and NGS (Illumina) Paired End sequencing BLAST Basic Local Alignment Search Tool Blast Program Selection Guide Table of Content 1. Introduction 2. BLAST Database Content 3. Program Selection Table 4. Explanation for the program choices given in Tables 3.1 and 3.2 5. Explanation for the program choices given in Tables 3.3 6. Explanation on Special Purpose Pages 7. Appendices 1
Basic Local Alignment Search Tool (BLAST) BLAST is a popular user-friendly tool for searching all the major sequence databases.BLAST is a heuristic method to find the highest scoring locally optimal alignments between a query sequence and a database sequence. BLAST programs BLAST Basic Local Alignment Search Tool Blast Program Selection Guide Table of Content 1. Introduction 2. BLAST Database Content 3. Program Selection Table 4. Explanation for the program choices given in Tables 3.1 and 3.2 5. Explanation for the program choices given in Tables 3.3 6. Explanation on Special Purpose Pages 7. Appendices 1
Basic Local Alignment Search Tool Stephen F. Altschul', Warren Gish', Webb Miller2 Eugene W. Myers3 and David J. Lipmanl 'Satwnal Center for Biotechnology Informalion Kdional Library of … BLAST (acronyme de basic local alignment search tool) est une méthode de recherche heuristique utilisée en bio-informatique. Il permet de trouver les régions similaires entre deux ou plusieurs séquences de nucléotides ou d'acides aminés, et de réaliser un alignement de ces régions homologues.
The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches. BLAST can be used to infer functional and evolutionary relationships between sequences as well as help identify Basic Local Alignment Search Tool Martin T otsches 14.06.2010 Martin T otsches Basic Local Alignment Search Tool. History Alignments Scores BLAST algorithm Conclusion Table of contents 1 History 2 Alignments global Alignments local Alignments 3 Scores Score functions Score matrices 4 BLAST algorithm Methods Statistical signi cance Performance 5 Conclusion Comparison to FASTA …
BLAST Basic Local Alignment Search Tool Job Title: lcl|P76387|WZC_ECOLI No comment found Show Conserved Domains Putative conserved domains have been … We have compared our result with the approximate solution obtained with BLAST (basic local alignment search tool) software, which is currently the most widely used for searching for protein alignment.
Basic Local Alignment Search Tool Stephen F. Altschul', Warren Gish', Webb Miller2 Eugene W. Myers3 and David J. Lipmanl 'Satwnal Center for Biotechnology Informalion Kdional Library of … 16/07/2016 · (2008) Blast (basic local alignment search tool). In: Encyclopedia of Genetics, Genomics, Proteomics and Informatics. Springer, Dordrecht.RIS Papers …
BLAST Basic Local Alignment Search Tool Blast Program Selection Guide Table of Content 1. Introduction 2. BLAST Database Content 3. Program Selection Table 4. Explanation for the program choices given in Tables 3.1 and 3.2 5. Explanation for the program choices given in Tables 3.3 6. Explanation on Special Purpose Pages 7. Appendices 1 Basic Local Alignment Search Tool Stephen F. Altschul', Warren Gish', Webb Miller2 Eugene W. Myers3 and David J. Lipmanl 'Satwnal Center for Biotechnology Informalion Kdional Library of …
The most common local alignment tool is BLAST (Basic Local Alignment Search Tool) developed by Altschul et al. (1990. J Mol Biol 215:403). The operative phrase in the phrase is local alignment. The BLAST is a set of algorithms that attempt to find a short fragment of a CSC 448 Bioinformatics Algorithms Alexander Dekhtyar. . (Rapid) Local Sequence Alignment BLAST BLAST: Basic Local Alignment Search Tool BLASTis afamilyof rapid approximate localalignmentalgorithms[2].
An Essential Guide to the Basic Local Alignment Search

Basic Local Alignment Search Tool(BLAST). This manual documents the BLAST (Basic Local Alignment Search Tool) command line applications developed at the National Center for Biotechnology Information (NCBI)., BLAST Basic Local Alignment Search Tool Job Title: lcl|P76387|WZC_ECOLI No comment found Show Conserved Domains Putative conserved domains have been ….
Jonathan M. Urbach Bioinformatics Group Department of
Basic Local Alignment Search Tool — Wikipédia. Created Date: 7/12/2001 9:55:01 AM https://ru.wikipedia.org/wiki/BLAST_(%D0%B7%D0%BD%D0%B0%D1%87%D0%B5%D0%BD%D0%B8%D1%8F) Wet lab Sampling (fish eye parasites) DNA extraction Primer design/ COI (marker ) PCR Library preparation and NGS (Illumina) Paired End sequencing.

BLAST - Basic Local Alignment Search Tool. Fonction. Alignement local d'une sГ©quence sur une banque de donnГ©es. Description . BLAST est un algorithme qui permet de trouver les rГ©gions similaires entre deux ou plusieurs sГ©quences de nuclГ©otides ou d'acides aminГ©s. Ce programme permet de calculer significativement les pourcentages de similitude entre les sГ©quences en les comparant avec (Basic Local Alignment Search Tool), which employs a measure based on well-defined mutation scores. It . directly approximates the results that would he obt,ained by a dynamic programming algoВ rithm for optimizing this measure. The method will detect weak but biologically significant sequence
BF children showed a significant enrichment in Bacteroidetes and depletion in Firmicutes (P < 0.001), with a unique abundance of bacteria from the genus Prevotella and Xylanibacter, known to contain a set of View Basic Local Alignment Search Tool.pdf from AA 1Basic Local Alignment Search Tool BLAST Why Use BLAST? BLAST helps you to find homologous genes and proteins Homologous Proteins (or genes) •
Recherche par similaritГ© dans les banques/bases de donnГ©es La suite Blast (Basic Local Alignment Search Tool) the Basic Local Alignment Search Tool Ian Korf, Mark Yandell & Joseph Bedell B LA ST Foreword by Stephen Altschul. BLAST. Other resources from OГ•Reilly oreilly.com oreilly.com is more than a complete catalog of O'Reilly books. You'll also find links to news, events, articles, weblogs, sample chapters, and code examples. oreillynet.com is the essential portal for developers interested in open
Basic Local Alignment Search Tool Stephen F. Altschul', Warren Gish', Webb Miller2 Eugene W. Myers3 and David J. Lipmanl 'Satwnal Center for Biotechnology Informalion Kdional Library of … The most common local alignment tool is BLAST (Basic Local Alignment Search Tool) developed by Altschul et al. (1990. J Mol Biol 215:403). The operative phrase in the phrase is local alignment. The BLAST is a set of algorithms that attempt to find a short fragment of a
search –Lower the E-value higher is the significance of score –P-value indicates if such an alignment can be expected from a chance alone Chance: can mean the comparison of (a) real but non-homologous sequences (True negatives) (b) real sequences that are shuffled to preserve compositional properties (c) sequences that are generated randomly search –Lower the E-value higher is the significance of score –P-value indicates if such an alignment can be expected from a chance alone Chance: can mean the comparison of (a) real but non-homologous sequences (True negatives) (b) real sequences that are shuffled to preserve compositional properties (c) sequences that are generated randomly
(Basic Local Alignment Search Tool), which employs a measure based on well-defined mutation scores. It . directly approximates the results that would he obt,ained by a dynamic programming algo rithm for optimizing this measure. The method will detect weak but biologically significant sequence View Basic Local Alignment Search Tool.pdf from AA 1Basic Local Alignment Search Tool BLAST Why Use BLAST? BLAST helps you to find homologous genes and proteins Homologous Proteins (or genes) •
(Basic Local Alignment Search Tool), which employs a measure based on well-defined mutation scores. It . directly approximates the results that would he obt,ained by a dynamic programming algo rithm for optimizing this measure. The method will detect weak but biologically significant sequence Introduction to Basic Local Alignment Search Tool. Chapter. 6 Readers; 1.3k Downloads; Summary . In this Chapter, we created a Swing based application that allows users to prepare sequences for BLAST searches by performing simple formatting tasks such as conversion into the Fasta format and determining the sequence type and length. Along the way we introduced how to write code to respond …
16/07/2016 · (2008) Blast (basic local alignment search tool). In: Encyclopedia of Genetics, Genomics, Proteomics and Informatics. Springer, Dordrecht.RIS Papers … 09/11/2017 · In this video, we describe the conceptual background and analysis method of Nucleotide BLAST (Basic Local Alignment Search Tool, BLASTn) analysis. Video on Local and Global Alignment …
1 BLAST: Basic Local Alignment Search Tool Jonathan M. Urbach Bioinformatics Group Department of Molecular Biology CSC 448 Bioinformatics Algorithms Alexander Dekhtyar. . (Rapid) Local Sequence Alignment BLAST BLAST: Basic Local Alignment Search Tool BLASTis afamilyof rapid approximate localalignmentalgorithms[2].
We have compared our result with the approximate solution obtained with BLAST (basic local alignment search tool) software, which is currently the most widely used for searching for protein alignment. BLAST Basic Local Alignment Search Tool Job Title: Borrprobe Your search parameters were adjusted to search for a short input sequence. BLASTN 2.2.17 (Aug-26-2007) Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, …
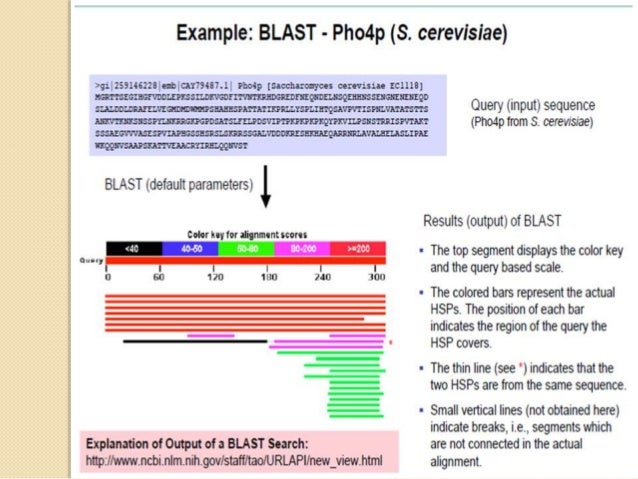
I The main strategy of BLAST is to search only those segment pairs that contain word with a score of at least threshold T. I Any such hit is extended to determine if it is contained within a segment pair whose score is greater than or equal to S. Sayyed Auwn Muhammad BLAST - Basic Local Alignment Search Tool I The main strategy of BLAST is to search only those segment pairs that contain word with a score of at least threshold T. I Any such hit is extended to determine if it is contained within a segment pair whose score is greater than or equal to S. Sayyed Auwn Muhammad BLAST - Basic Local Alignment Search Tool